Lecture Materials
Ising model#
Ising model represents a system of spins (magnetic dipoles) on a lattice. Without external magnetic field, the energy reads
where \(J > 0\) for a ferromagnetic. For nearest-neighbor interaction only, the sum runs over the pairs of neighboring lattice sites.
Here each dipole interacts with its four neighbors (or two/three neighbors in case of the spins at the edges). The magnetization is given by
It is know that below the Curie temperature
the system attains a spontaneous magnetization \(|M| > 0\). We can simulate this process using Markov chain and the Metropolis algorithm.
Let us apply the Metropolis algorithm to simulate the system. Each state is given by the spin configuration \(\{ s_i \}\). At each step we randomly choose one spin \(i\) and flip its orientation \(s_i\) to \(-s_i\). Such a flip would cause the energy of the system to change by
The new state is then accepted with a probability
import numpy as np
import matplotlib.pyplot as plt
# Energy of 2D Ising system for a given spin configuration
def IsingE(spins, periodicBC = False):
energy = 0
N = len(spins)
for i in range(N):
for j in range(N):
if (i > 0 or periodicBC):
energy += -spins[i][j] * spins[i-1][j]
if (j > 0 or periodicBC):
energy += -spins[i][j] * spins[i][j-1]
if (i < N - 1 or periodicBC):
energy += -spins[i][j] * spins[(i+1)%N][j]
if (j < N - 1 or periodicBC):
energy += -spins[i][j] * spins[i][(j+1)%N]
return energy / 2
# Magnetization of 2D Ising system for a given spin configuration
def IsingM(spins):
return np.sum(spins)
# Change of energy of the Ising system by flipping the spin at the location (i,j)
def IsingdEflip(spins, i, j, periodicBC = False):
N = len(spins)
dE = 0
if (i > 0 or periodicBC):
dE += 2 * spins[i][j] * spins[i-1][j]
if (j > 0 or periodicBC):
dE += 2 * spins[i][j] * spins[i][j-1]
if (i < N - 1 or periodicBC):
dE += 2 * spins[i][j] * spins[(i+1)%N][j]
if (j < N - 1 or periodicBC):
dE += 2 * spins[i][j] * spins[i][(j+1)%N]
return dE
from IPython.display import clear_output
from matplotlib.colors import ListedColormap
def iterateIsing(spins, T, N, steps, periodicBC = False):
eplot = []
Mplot = []
E = IsingE(spins, periodicBC)
M = IsingM(spins)
eplot.append(E)
Mplot.append(M)
for k in range(steps):
# Pick the lattice site randomly
i = np.random.randint(N)
j = np.random.randint(N)
# Energy change from flipping the site
dE = IsingdEflip(spins, i, j, periodicBC)
# Flip the spin with some probability
if (np.random.rand() < np.exp(-dE/T)):
spins[i,j] = -spins[i,j]
E += dE
M += 2 * spins[i,j]
eplot.append(E)
Mplot.append(M)
return eplot, Mplot
# Simulates the 2D Ising system of NxN spins at temperature T
# by performing Markov chain steps using Metropolis algorithm
# Returns arrays energies and magnetizations at each step
def simulateIsing(T, N, steps, periodicBC = False):
spins = -1 + 2 * np.random.randint(0, high = 2, size=(N,N))
eplot, Mplot = iterateIsing(spins, T, N, steps, periodicBC)
return eplot, Mplot
Let us simulate the system at \(T = 1 < T_C\) several times and keep track of the magnetization.
%%time
N = 20
steps = 500000
periodicBC = True
Temperature = 1. # Tc = 2. / np.log(1+np.sqrt(2.)) \approx 2.27
Ts = np.empty(5)
Ts.fill(Temperature)
eplots = []
Mplots = []
plotSimulation = False
for T in Ts:
resE, resM = simulateIsing(T, N, steps, periodicBC)
eplots.append(resE)
Mplots.append(resM)
# Make the graph
import matplotlib.pyplot as plt
for i in range(len(Ts)):
leg = "Run " + str(i + 1)
plt.plot(Mplots[i],label=leg)
plt.title("2D Ising model, T = " + str(Temperature))
plt.xlabel("Step")
plt.ylabel("Magnetisation")
plt.legend()
plt.show()
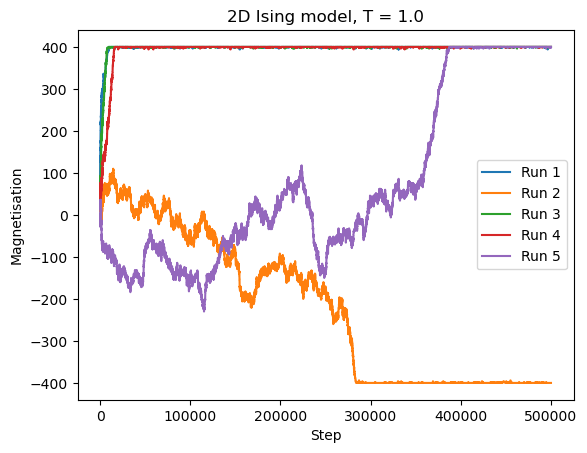
CPU times: user 12.6 s, sys: 106 ms, total: 12.7 s
Wall time: 12.3 s
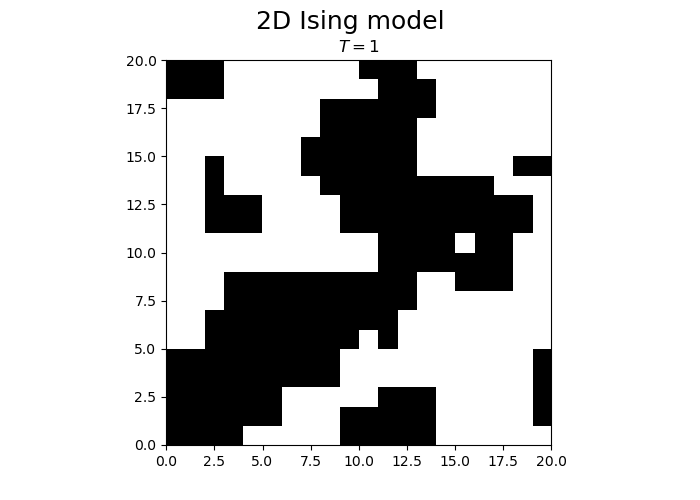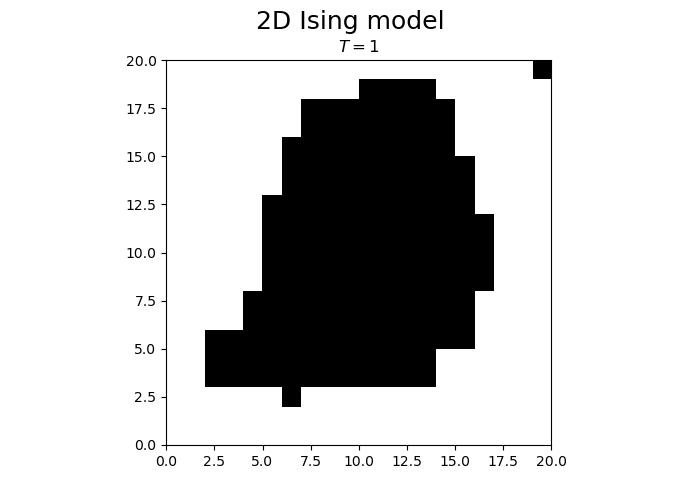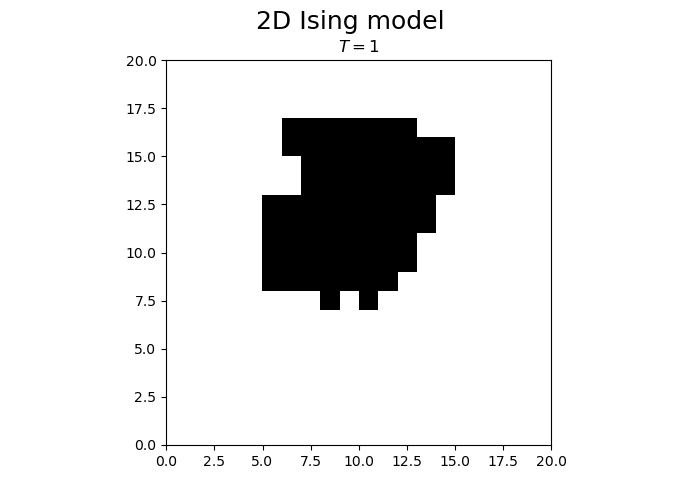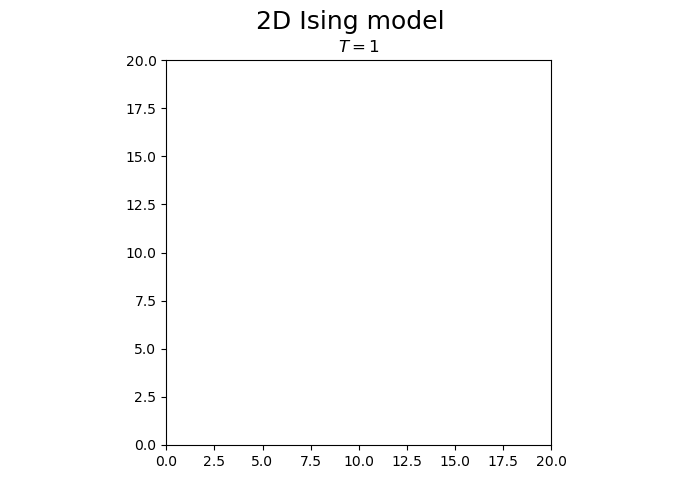
N = 20
T = 1
steps = N*N
spins = -1 + 2 * np.random.randint(0, high = 2, size=(N,N))
from matplotlib.animation import FuncAnimation
fig, ax = plt.subplots(1, 1)
fig.set_size_inches(7, 5, forward=True)
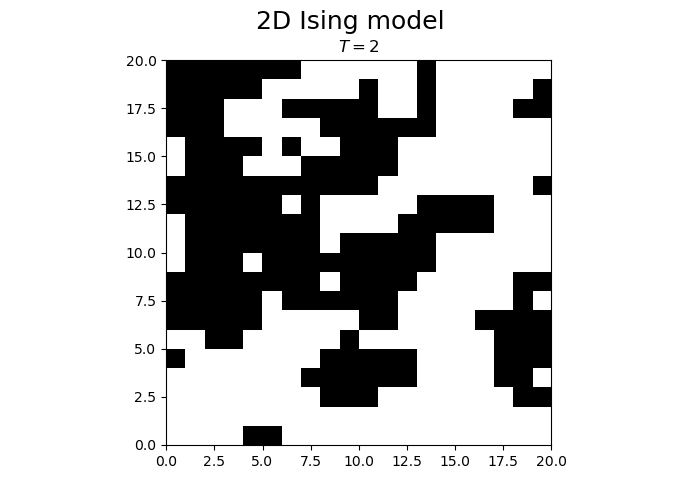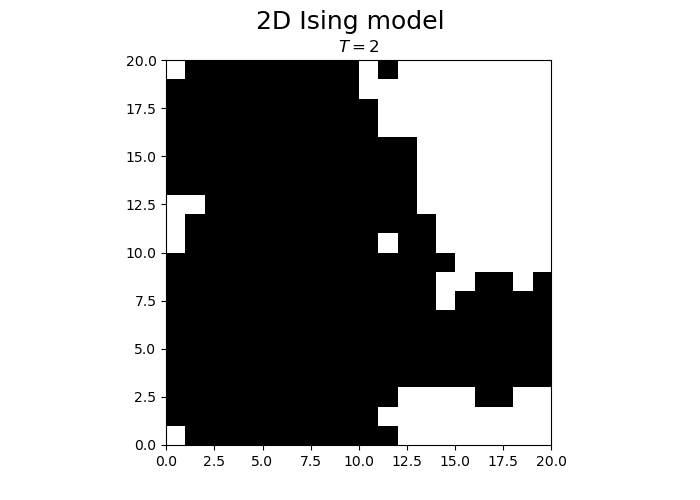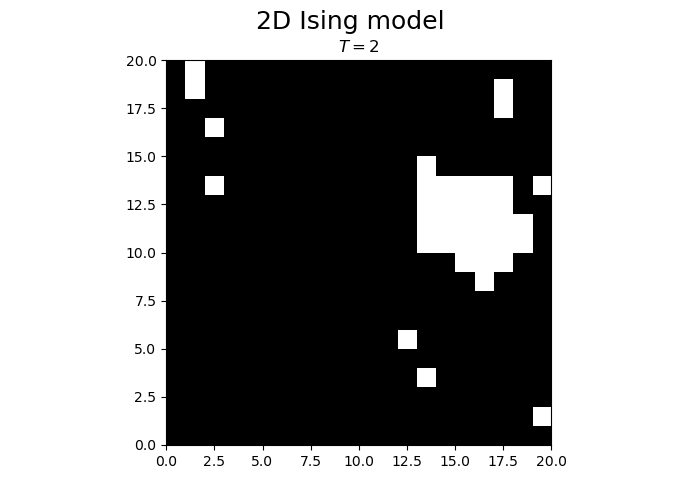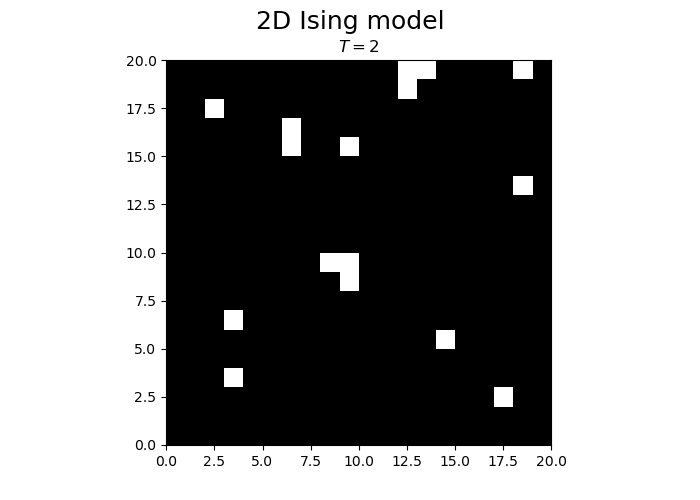
fig.suptitle("2D Ising model", fontsize = 18)
im = ax.imshow(spins, cmap='gray', vmax=1, vmin=-1, origin="lower", extent=[0,N,0,N])
title = ax.set_title("")
def animateIsing(i):
iterateIsing(spins, T, N, N*N, periodicBC = True)
im.set_data(spins)
title.set_text("$T = $" + str(T))
return im, title
ani = FuncAnimation(fig, animateIsing, frames=30*3, interval=50, blit=True)
plt.close()
ani.save("ising.gif")
# from IPython.display import HTML
# HTML(ani.to_jshtml())
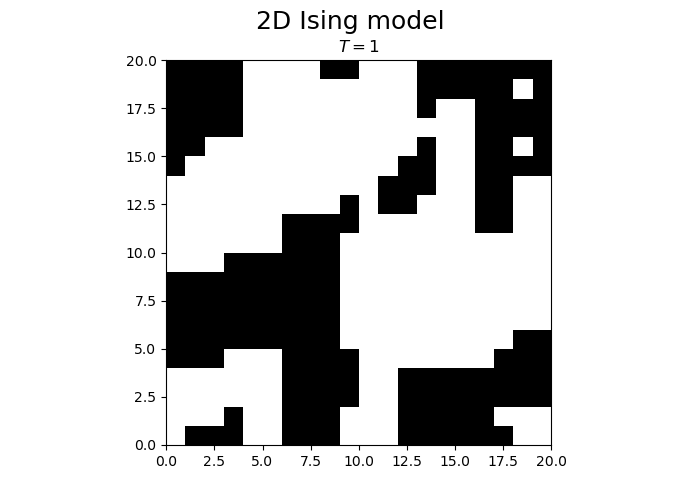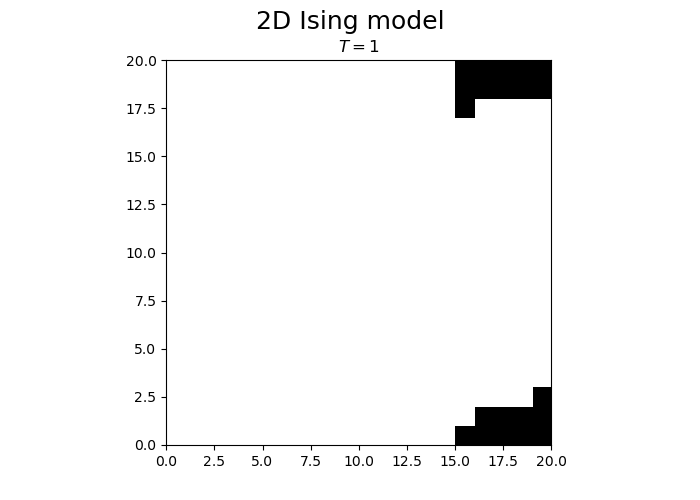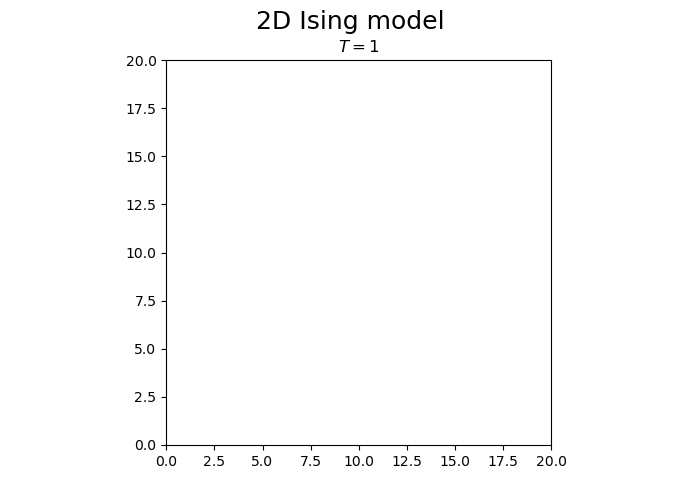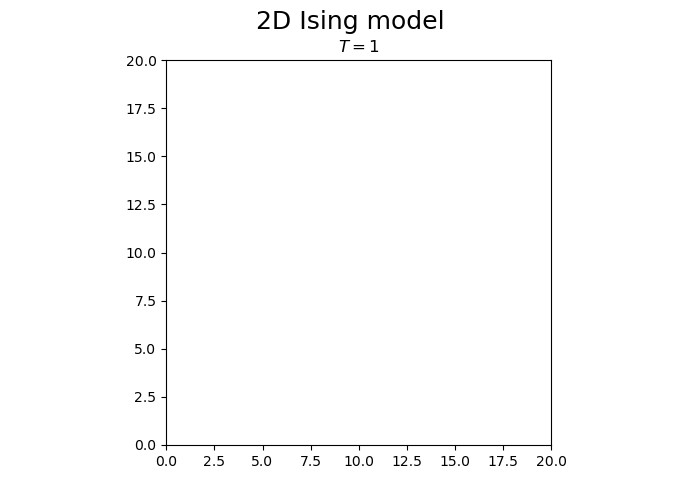
We observe that the magnetization spontaneously attains either positive or negative values.
Let us now consider different temperatures.
%%time
N = 20
steps = 1000000
periodicBC = True
Ts = [1., 2., 3.]
eplots = []
Mplots = []
plotSimulation = False
for T in Ts:
resE, resM = simulateIsing(T, N, steps, periodicBC)
eplots.append(resE)
Mplots.append(resM)
# Make the graph
import matplotlib.pyplot as plt
for i in range(len(Ts)):
leg = "T = " + str(Ts[i])
plt.plot(Mplots[i],label=leg)
plt.title("2D Ising model")
plt.xlabel("Step")
plt.ylabel("Magnetisation")
plt.legend()
plt.show()
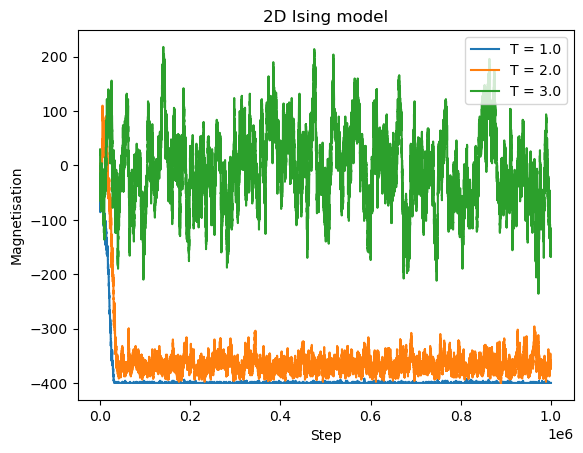
CPU times: user 18.5 s, sys: 2.83 s, total: 21.3 s
Wall time: 18.4 s
The system reaches non-zero magnetization below the Curie temperature \(T_C \approx 2.27\), and the process is faster at lower temperatures. For \(T = 3 > T_C\), the magnetization fluctuates around zero.
|
|
|
|---|