Lecture Materials
Boundary value problems#
Laplace’s equation#
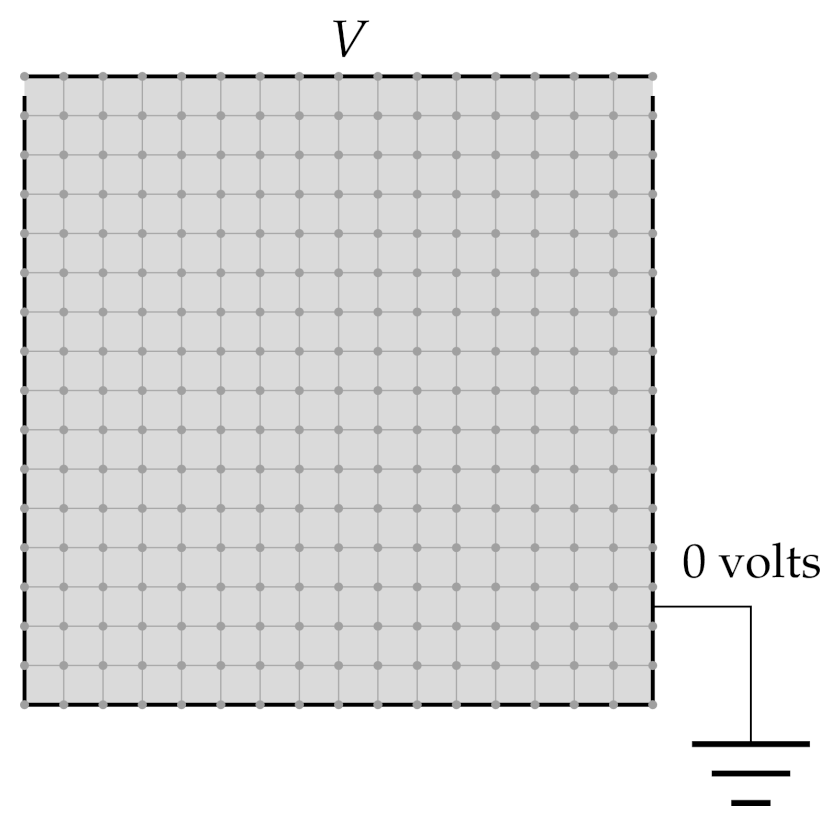
Boundary value problems typically deal with static solutions (no time variable)
Consider Laplace’s equation (no external charges) in two dimensions
Boundary conditions: one of the walls has voltage \(V\) applied to it, the rest are grounded:
Let us apply finite difference method. Discretize the box into a grid in steps \(a = L / M\) in each direction. Approximate the derivatives by central differences:
Laplace’s equation becomes
In principle, this is a system of \(N \sim M^2\) linear equations that can be solved exactly. The general method will however require \(O(N^3) = O(M^6)\) operations which becomes unfeasible already for \(M \sim 100\) or so.
Jacobi (relaxation) method#
Start with some initial guess \(\phi_0(x,y)\). The next iteration is given by
for all points inside the box. Preserve the boundary conditions at each iteration:
This is the Jacobi (or relaxation) method, and for Laplace’s equation it is always converges. Let us implement it in Python:
import numpy as np
# Single iteration of the Jacobi method
# The new field is written into phinew
def iteration_jacobi(phinew, phi):
M = len(phi) - 1
# Boundary conditions
phinew[0,:] = phi[0,:]
phinew[M,:] = phi[M,:]
phinew[:,0] = phi[:,0]
phinew[:,M] = phi[:,M]
for i in range(1,M):
for j in range(1,M):
phinew[i,j] = (phi[i+1,j] + phi[i-1,j] + phi[i,j+1] + phi[i,j-1])/4
delta = np.max(abs(phi-phinew))
return delta
def jacobi_solve(phi0, target_accuracy = 1e-6, max_iterations = 100):
delta = target_accuracy + 1.
phi = phi0.copy()
for i in range(max_iterations):
delta = iteration_jacobi(phi, phi0)
phi0, phi = phi, phi0
if (delta <= target_accuracy):
print("Jacobi method converged in " + str(i+1) + " iterations")
return phi0
print("Jacobi method failed to converge to a required precision in " + str(max_iterations) + " iterations")
print("The error estimate is ", delta)
return phi
%%time
# Measure execution time
# Constants
M = 100 # Grid squares on a side
V = 1.0 # Voltage at top wall
target = 1e-4 # Target accuracy
# Initialize with zeros
phi = np.zeros([M+1,M+1],float)
# Boundary condition
phi[0,:] = V
phi[:,0] = 0
phi = jacobi_solve(phi, target, 10000)
# Plot
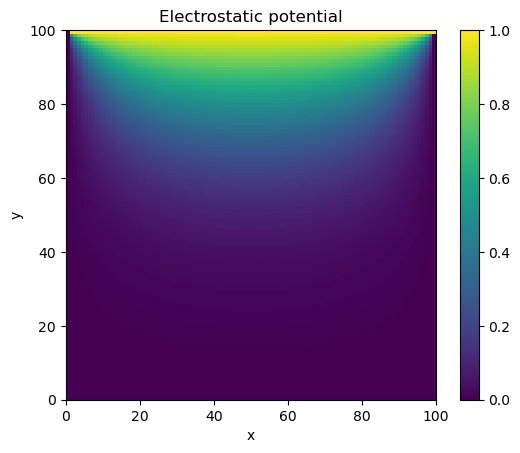
import matplotlib.pyplot as plt
plt.title("Electrostatic potential")
plt.xlabel("x")
plt.ylabel("y")
CS = plt.imshow(phi, vmax=1., vmin=0.,origin="upper",extent=[0,M,0,M])
plt.colorbar(CS)
plt.show()
Jacobi method converged in 1909 iterations
CPU times: user 8.95 s, sys: 845 ms, total: 9.79 s
Wall time: 8.55 s
Gauss-Seidel method with overrelaxation#
The base Jacobi method corresponds to an iteration
This requires to have two arrays independently. In Gauss-Seidel method one uses the already computed values of \(\phi_{n+1}\) where availabl instead. The method thus corresponds to
Another modification is the use of overrelaxation.
The base Jacobi method is a type of relaxation method
A way to speed-up calculation is to overrelaxate the solution a bit
where \(\omega > 0\).
This implies the following iterative procedure
This is unstable for \(\omega > 0\). However, applied to Gauss-Seidel method, this corresponds to
and provides generally stable solution for \(\omega < 1\).
import numpy as np
# Single iteration of the Gauss-Seidel method
# The new field is written into phi directly
# omega >= 0 is the overrelaxation parameter
def gaussseidel_iteration(phi, omega = 0):
M = len(phi) - 1
delta = 0.
# New iteration
for i in range(1,M):
for j in range(1,M):
phiold = phi[i,j]
phi[i,j] = (1. + omega) * (phi[i+1,j] + phi[i-1,j] + phi[i,j+1] + phi[i,j-1])/4 - omega * phi[i,j]
delta = np.maximum(delta, abs(phiold - phi[i,j]))
return delta
def gaussseidel_solve(phi0, omega = 0, target_accuracy = 1e-6, max_iterations = 100):
delta = target_accuracy + 1.
phi = phi0.copy()
for i in range(max_iterations):
delta = gaussseidel_iteration(phi, omega)
if (delta <= target_accuracy):
print("Gauss-Seidel method converged in " + str(i+1) + " iterations")
return phi
print("Gauss-Seidel method failed to converge to a required precision in " + str(max_iterations) + " iterations")
print("The error estimate is ", delta)
return phi
Let us check the performance of the Gauss-Seidel method with overrelaxation.
%%time
# Constants
M = 100 # Grid squares on a side
V = 1.0 # Voltage at top wall
target = 1e-4 # Target accuracy
# Initialize with zeros
phi = np.zeros([M+1,M+1],float)
# Boundary condition
phi[0,:] = V
phi[:,0] = 0
omega = 0.93
phi = gaussseidel_solve(phi, omega, target, 1000)
# Plot
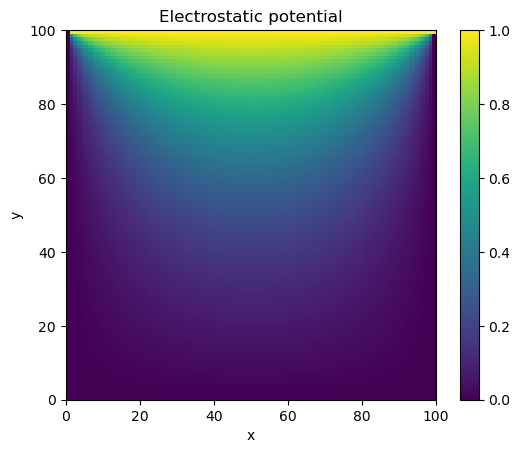
plt.title("Electrostatic potential")
plt.xlabel("x")
plt.ylabel("y")
CS = plt.imshow(phi, vmax=1., vmin=0.,origin="upper",extent=[0,M,0,M])
plt.colorbar(CS)
plt.show()
Gauss-Seidel method converged in 137 iterations
CPU times: user 2.33 s, sys: 4.67 ms, total: 2.33 s
Wall time: 1.84 s

